Optimal Leaf Ordering
Heirarchial Clustering and UPGMA
Hierarchical clustering clusters a set of input elements into a
binary tree with similar groups clustered together. Hierarchial clustering
is useful in organizing large sample of data, and has been used
extensively, amongst others, in biological study of data samples. The distance
between the two groups clustered together is based in terms of the
similarities between the groups. For instance, for a given sample of DNA sequences,
two genes with more CG content will be grouped together because they are more similar.
In this program, we use the UPGMA method of hierarchical clustering. UPGMA, acronym for
Unweighted Pair Grouping with Arithmetic Mean, is an algorithm used in bioinformatics
for the creation of a dendrogram (i.e. a rooted tree), with the assumption that
there is a constant rate of evolution (between the gene samples from orthologs/paralogs,
for instance). For more information about the UPGMA algorithm, please refer to this
wikipedia site.
Optimal Ordering
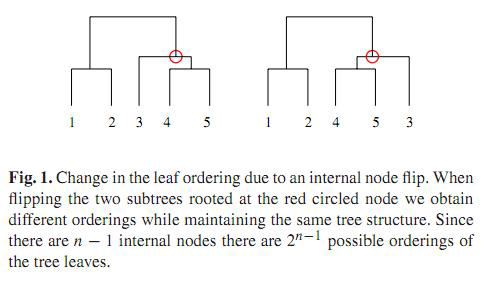 A UPGMA dendrogram with any one of its internal node flipped will yield different ordering of the leaves,
while the tree structure remains the same. Since biological analysis is often done in the context of linear
arrangement of the leaves themselves, the linear ordering of the leaves become significant.
A UPGMA dendrogram with any one of its internal node flipped will yield different ordering of the leaves,
while the tree structure remains the same. Since biological analysis is often done in the context of linear
arrangement of the leaves themselves, the linear ordering of the leaves become significant.
The Optimal Leaf Ordering attempts to cluster similar groups (or leaves) taken from the UPGMA algorithm, and yields the leaf order that maximizes the sum of the similarities of adjacent leaves in the ordering. For the Optimal Leaf Ordering algorithm, please refer to the paper "Fast Optimal Leaf Ordering for Heirarchical Clustering" by Ziv B. J. et. al. (published in Bioinformatics 17 2001 S22-S29). More information about the algorithm can be found here.