ENTERING DATA
There are two methods by which data can be entered for analysis, matrix and sequence. The matrix option allows you
to enter your own distance values with labels for each taxon. This method should be used when have already used some method of
distance calculation to determine the distances between taxa. A line should be entered for each row in the distance matrix,
beginning with the name you wish to assign to each taxon, as shown:
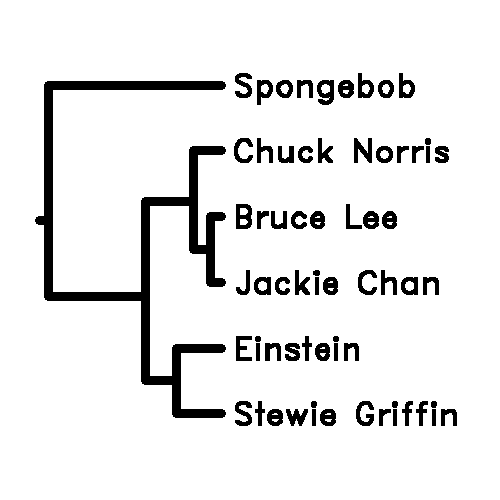
Einstein 0 4 6 9 12 5
Stewie_Griffin 4 0 7 8 10 2
Bruce_Lee 6 7 0 2 17 1
Chuck_Norris 9 8 2 0 28 3
Spongebob 12 10 17 28 0 6
Jackie_Chan 5 2 1 3 6 0
In order to ensure that the program can access the data, be sure that the names of your taxa (far left colum) do not contain spaces.
Use underscores instead. Also make sure, however, that you place spaces between each entry and make a separate line for each taxon.
Alternatively, you can enter a set of DNA sequences of equal length and use these to build a tree. For each sequence, list the name
that you wish to give that taxon followed by a semicolon and then the sequence, as shown below. each sequence should go on a separate
line.
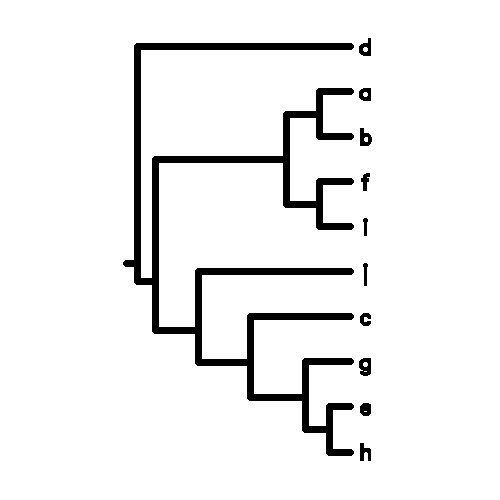
a:GGATAGAGAC
b:GGAGAGTTCC
c:GAGAGAAACT
d:TTCTCTCTAG
e:GATTAGACAA
f:TTAGAGTAGT
g:TAGTAGCAGA
h:GATTAAGAAA
i:GGAGATTAGA
j:CCTATCAGCA
Putting spaces before or after the ":" is alright (a : GATTACA), but there should be no spaces between the letters in your sequence,
(a : G ATT A CA) will not work.