

The purpose of this prediction program is to find the most likely locations along an amino acid sequence for protein antigenic determinants. In this method, each amino acid is assigned a hydrophility value and then these values are repetitively averaged along the inputted amino acid chain.

Antigens
First off, what is a protein antigenic determinant and why do we want to locate them? A protein antigen elicits the formation of antibodies once introduced to the foreign individual. These antibodies form according to the number of antigenic determinants in the protein's amino acid sequence as demonstrated in the diagram below.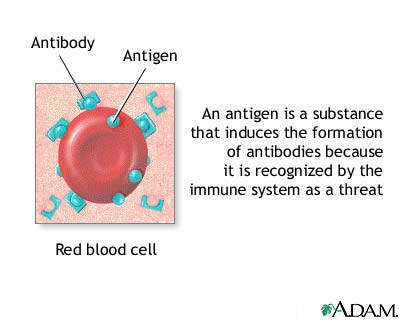
(Source)
So when an individual is immunized against some sort of disease, what's really happening is that a protein antigen is being introduced to the body and the body forms new antibody molecules with different amino acid specificities. Antigens can also be produced within the individual's own cells. Antigens may be excreted from the cell or released after cell death. These antigens often turn out to be toxic and the presence of them within an animal is evidence of past or present contact with certain microorganisms. Therefore, it is valuable to predict where antigenic determinants are located on a particular protein because this knowledge would be beneficial to seeing just how prepared a cell is to foreign antigen attack and also to see if it would be possible to predict if the cell containing such a protein would pose a threat to itself. However it should be noted that not all proteins contain antigenic determinants.
Information researched on Answers.com.

The Hopp and Wood protein antigen determinant prediction method
This program seeks to identify protein antigen determinants through hydrophilicity maxima. Hopp and Wood correlated the antigenic sites with the amino acid's hydrophilicity because the relationship was supported in a few test cases of known antigen proteins. It is assumed that most antigen determinant sites are exposed on the outside folds of the protein, and therefore must be tolerant of a hydrophilic environment. So the higher the average hydrophilicity value of a group (window) of amino acids, the more likely it would be to code for an antigen. After testing different windows on a well known antigen-coding protein, Hopp and Wood determined that a window size of 6 yielded the best and most accurate results as to the locations of the antigen determinants. These locations are best visualized on a plot of average hydrophilicity value versus sequence position. The positive peaks in the curve represent possible protein antigen determinants!
The Hopp and Wood paper can be found here.
